Here you find documents such as policies, presentations, posters and flyers produced by Solve-RD.
Results

Solve-RD Data Sharing Policy
The Solve-RD Data Sharing Policy explains how data is being shared witin Solve-RD and outside the project (e.g. via the RD-Connect Genome-Phenome Analysis Platform). The most recent version (version 8) has been accepted by the General Assembly on 24 January 2022.

Solve-RD Publication Policy
The Solve-RD Publication Policy provides detailed information about authors' responsibilities with regard to publication of shared data and data resulting from the Solve-RD project. It also explains how to acknowledge Solve-RD funding, use of the RD-Connect Genome-Phenome Analysis Platform and ERN involvement.

Code of Conduct
The Solve-RD Code of Conduct has been generated to regulate the use of the Solve-RD analysis sandbox and access to Solve-RD data. All Solve-RD investigators requesting access to the sandbox and to Solve-RD data need to sign the Code of Conduct.
The template to submit analysis/usage projects can be downloaded here.
Video: Solve-RD webinar on the new graphical user interface of the RD-Connect GPAP
The Solve-RD webinar on the new graphical user interface of the RD-Connect Genome-Phenome Analysis Platform (GPAP) given by Leslie Matalonga (CNAG-CRG) on 12 April 2023 is now available online via this link!
The webinar focuses on how to navigate through the platform and use some of the advanced new functionalities to (re)analyse cases, including cohort level analysis and the use of Matchmaker Exchange.
Video: Solve-RD webinar on advanced features of the RD-Connect GPAP
Video: Solve-RD webinar on basic analysis using the RD-Connect GPAP

Treatabolome
Solve-RD WP3 aims at changing patient lives via the establishment of a "Treatabolome" database flagging treatable genes and variants which will contribute to the translation of genomics data into the clinical setting.
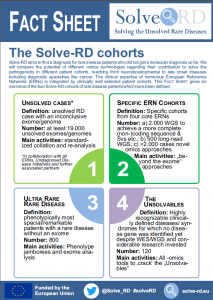
Fact Sheet: Solve-RD cohorts
Solve-RD aims to find a diagnosis for rare disease patients who did not get a molecular diagnosis so far. We will compare the potential of different -omics technologies regarding their contribution to solve the pathogenicity in different patient cohorts, reaching from neurodevelopmental to late onset diseases including diagnostic specialties like cancer. The clinical expertise of numerous European Reference Networks (ERNs) is integrated by clinically well selected patient cohorts. This FACT SHEET gives an overview of the four Solve-RD cohorts of rare disease patients which have been defined.

Solve-RD – Solving the Unsolved Rare Diseases.
A few slides to get an impression of the project.

Solve-RD poster
The Solve-RD poster has been presented at the ECRD conference in Vienna 2018 and at the ESHG conference in Milan 2018.
