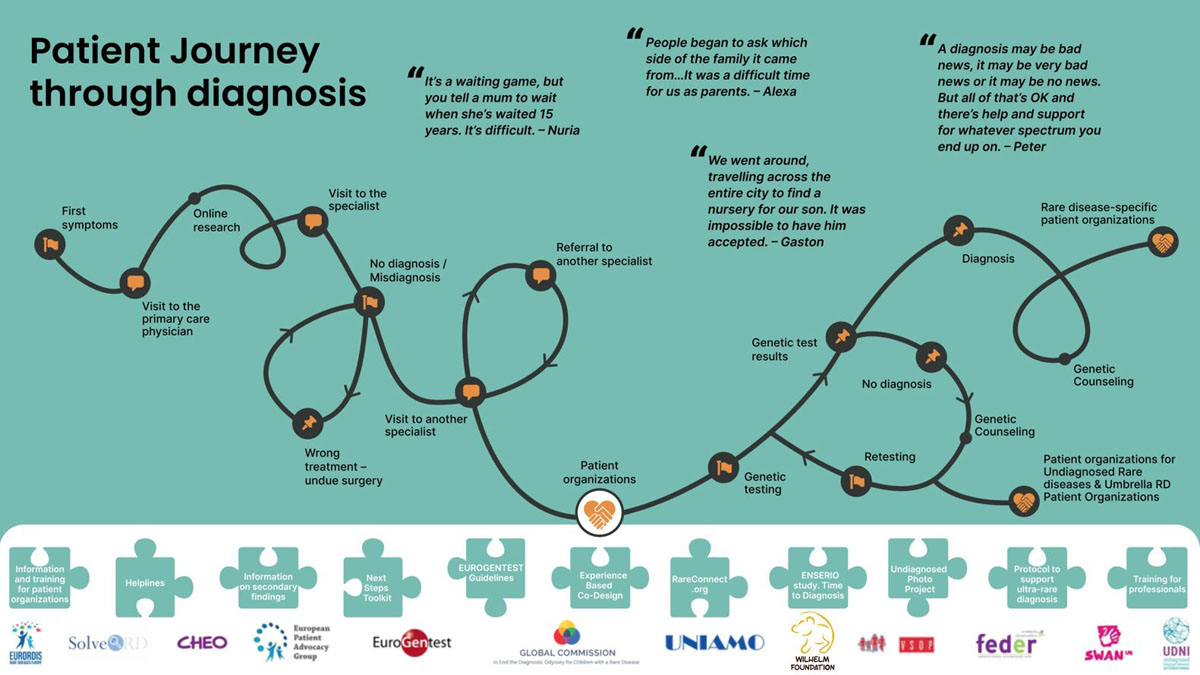
Diagnostic odyssey
The EURORDIS-led Community Engagement Task Force (CETF) has created an infographic setting out the patient journey to diagnosis. The infographic demonstrates the diagnostic odyssey many people experience on a daily basis and presents existing resources from CETF member organisations to support patients on this journey. The infographic is available in 16 languages!
You will find more info here.




